How second order random walk on graph works, explained via animations

Node2vec is an embedding method that transforms graphs (or networks) into numerical representations [1]. For example, given a social network where people (nodes) interact via relations (edges), node2vec generates numerical representation, i.e., a list of numbers, to represent each person. This representation preserves the structure of the original network in some sense so that people closely related have similar representations and vice versa. In this article, we’ll go through the intuition of the node2vec method and, in particular, how second order random walk on graph works via a series of animations.
TL;DR
- Node2vec generates numerical representations of nodes in the graph via 2nd order (biased) random walk.
- First order random walk is done by sampling nodes on the graph along the edges of the graph, and each step depends only on the current state.
- Second order random walk is a modified version of the first order random walk that depends not only on the current state but also the previous state.
- A corpus of random walks is generated using each node in the network as a starting point. This corpus is then fed through word2vec to generate final node embeddings.
First order random walk
A random walk on a graph can be thought of as a "walker" traversing the graph along the edges of the graph. At each step, the walker needs to decide where to travel next and then move to the next state. This process is referred to as a 1-hop transition. Let’s take a look at a simple example.

In this example, let’s assume the walker is currently on v, which has three neighboring nodes: u1, u2, and u3. Each neighbor connects to v with different weights, as shown in the figure. These weights are used to determine the probability of picking the next step. Specifically, the transition probabilities __ are calculated by normalizing the edge weights:

where N_v is the set of neighboring nodes of v, which are u1, u2, and u3 in this case, and d(v) is the degree of node v.
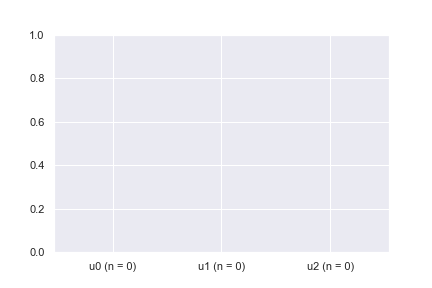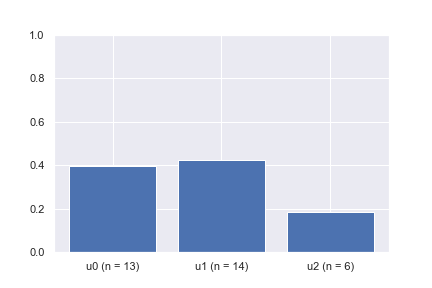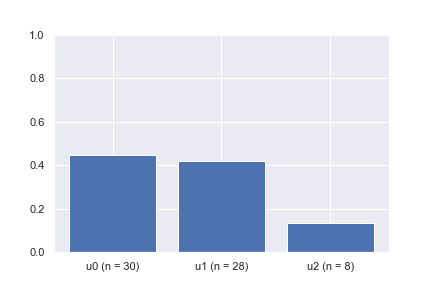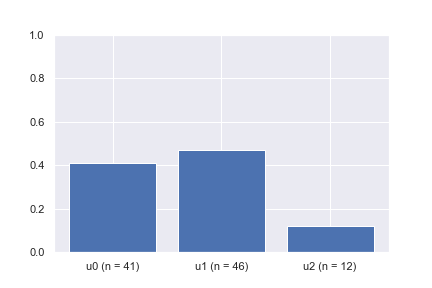
After a quick calculation, we see that the transition probabilities from v to u1, u2, and u3 are 0.4, 0.5, and 0.1, respectively. By the law of large numbers, we know that if we draw enough samples (i.e., pick the next state given the current state v) using the specified transition probabilities, the average transitions should be close to the expected values. The bar plot animation shows the example of picking the next state given the current state at v a hundred times. It can be seen that the percentage of times each neighboring node is picked is converging to the desired probabilities of 0.4, 0.5, and 0.1, respectively.
Now that we understand how 1-hop transition is made via transition probabilities, we can move on to random walk generation. In short, a random walk is generated by carrying out multiple 1-hop transitions, where the previous 1-hop transition determines the current state.
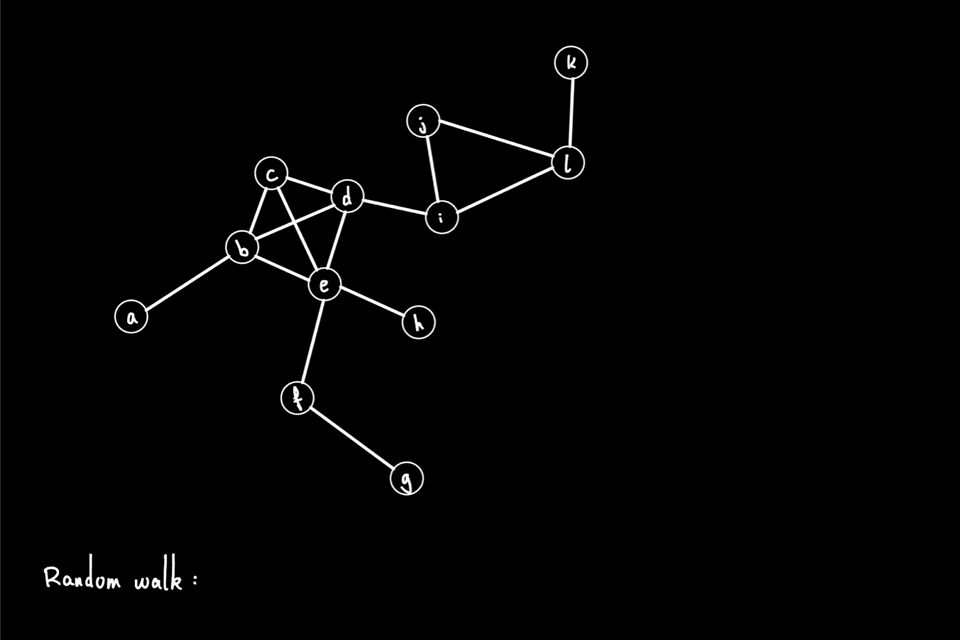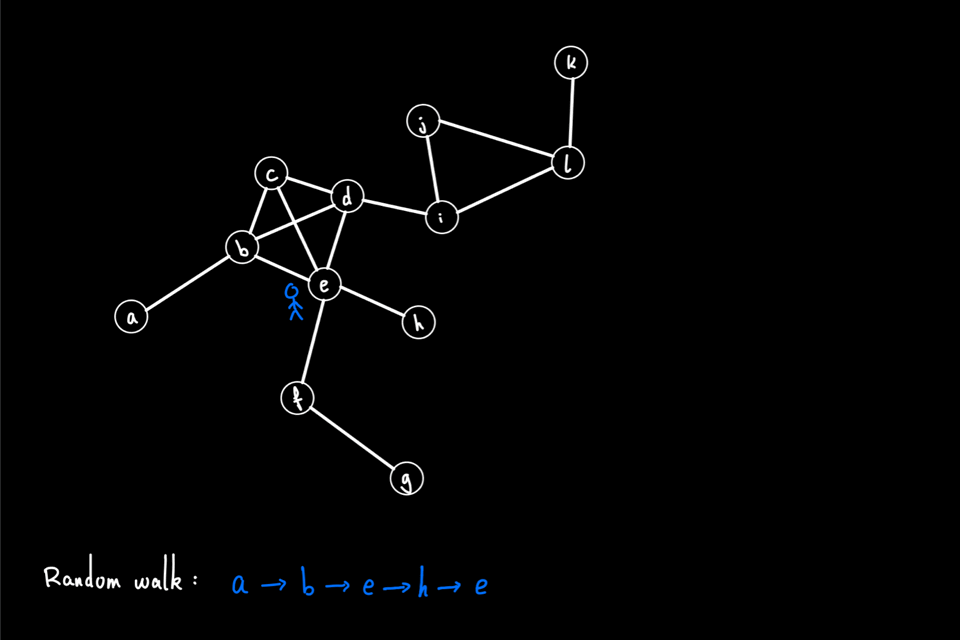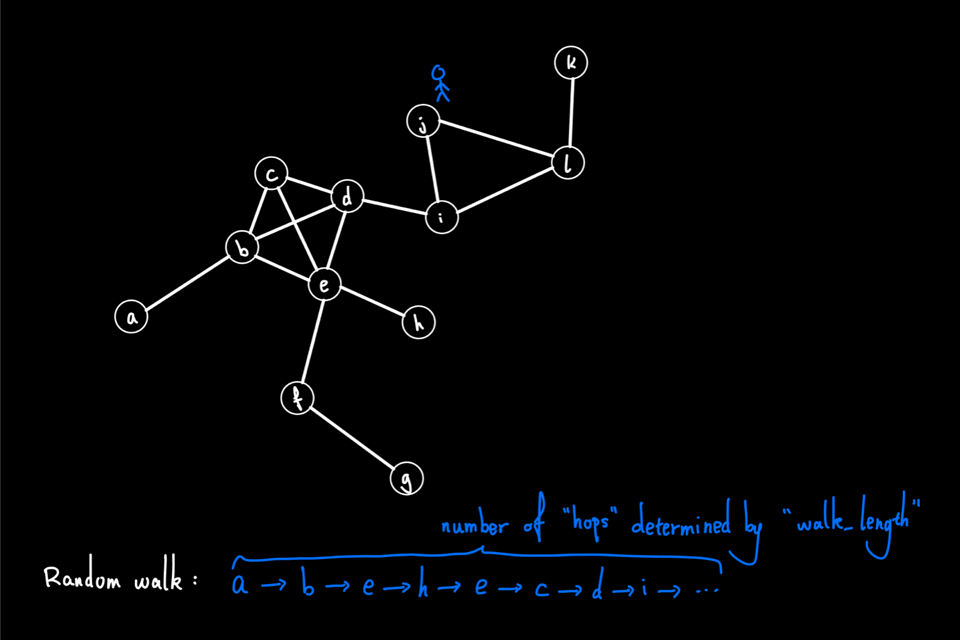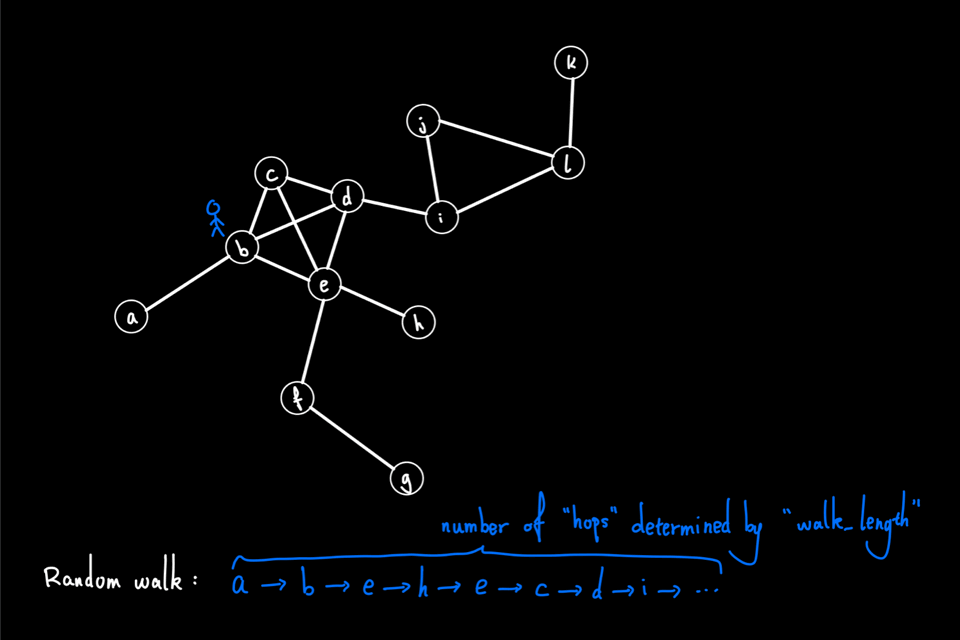
The above animation shows an example of a random walk on a graph with 12 nodes (node "a" through node "l"), starting from node "a". In each iteration, the walker proceeds to the next node via a 1-hop transition. The random walk process continues until either it reaches the predefined "walk_length" or the walker reaches a "dead end" for a directed graph, i.e., the node has no outgoing edge. In this example, the random walk generated is the node sequence: [a, b, e, h, e, c, d, i, …]
What we’ve seen so far is called the first order random walk, which means that the 1-hop transition depends only on the current state. However, node2vec performs second order random walk, which is a slightly modified version that incorporates information from the previous step.
Second order biased walk
Instead of looking at direct neighbors of the current state, the second order transition applies a bias factor alpha to reweigh the edge weights depending on the previous state. In particular, alpha is a function that takes as inputs the current state and the potential next state. In the first case, if two states (nodes) are not connected, then alpha is set to be 1/q, where q is the in-out parameter. Thus one can either increase the probability of going "outward" (meaning that the walk is not restricted in a localized neighborhood) by specifying a small q value or conversely restrict the walks to a local neighborhood by setting a large q value. In the second case, if the two states are identical, meaning that the walk is "returning" back to the previous state, then alpha is set to be 1/p, where p is the return parameter. Finally, if the two states are not identical and are connected, then alpha is set to 1.
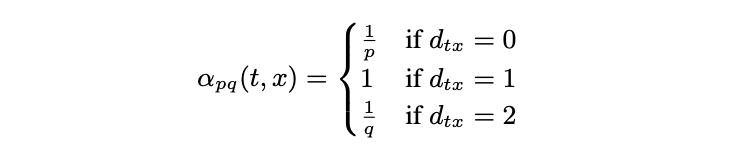
Returning to our previous example on 1-hop transition and assuming that u2 is the previous state, the figure below highlighted the return edge with red and in-out edge with yellow. The bias factors alpha are assigned accordingly: 1/q for in-out (yellow), 1/p for return (red), and 1 otherwise.

With these bias factors in hand, we can proceed to calculate the 2nd order transition probabilities as follow, which is very similar to what we’ve seen above for 1st order transition, but now each weight is biased by alpha.

As before, the 2nd order random walk can be generated using the 2nd order transition probabilities. In the animation below, the return and in-out edges are highlighted with red and yellow, respectively.

Node embeddings from random walks
So far, the random walks we’ve seen are initiated from a single node. Imagine that this random walk generation process is repeated multiple times using each node in the graph as the initial node. Then, as a result, we have a large "corpus" of node sequences. This "corpus" can then be directly fed into the word2vec algorithm to generate node embeddings. In particular, node2vec uses skipgram with negative sampling (short for SGNS). The details of SGNS are omitted here, but I strongly encourage readers to check out two great blog posts ([2] and [3]) that explained SG and NS, respectively, by Chris McCormic.
The animation below gives a brief idea of training the skipgram using the random walks generated on the graph. The main idea is to maximize the probability (calculated via softmax) of predicting the correct context node given center node. On the other hand, negative sampling is used to improve computational efficiency by only computing a couple of the activation from randomly drawn "negative samples" instead of the full normalization factor in the softmax.
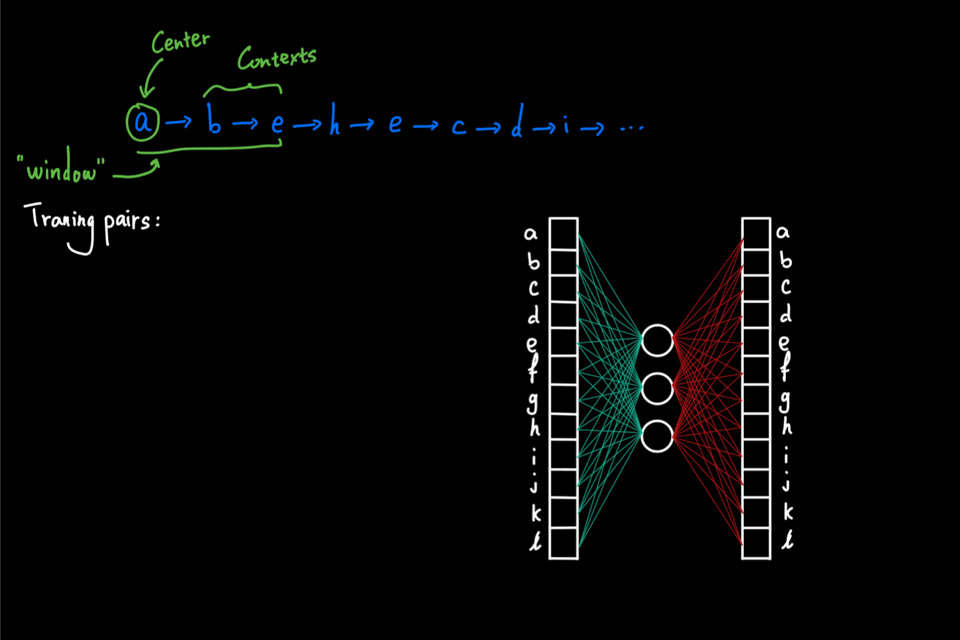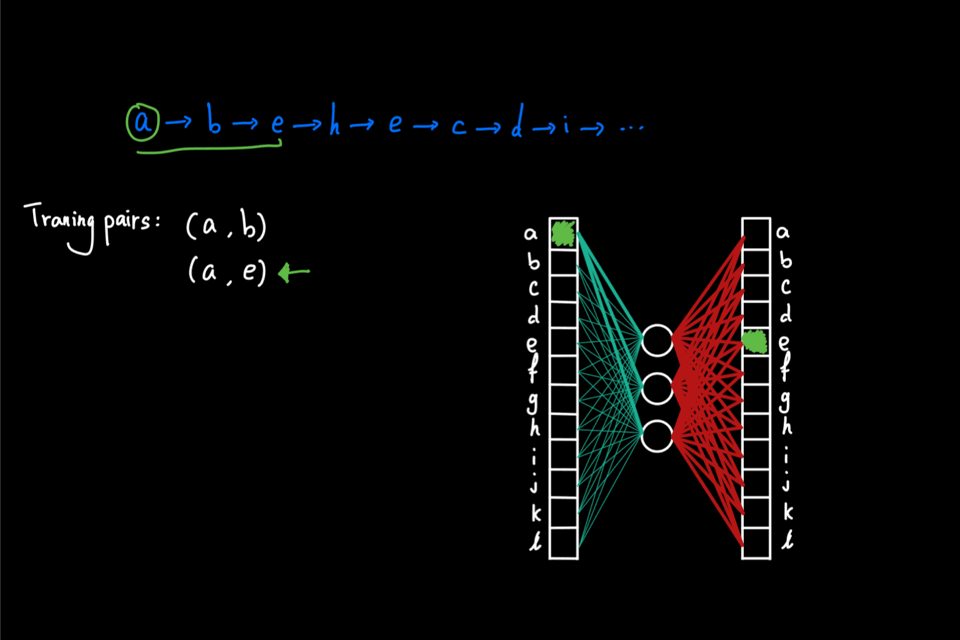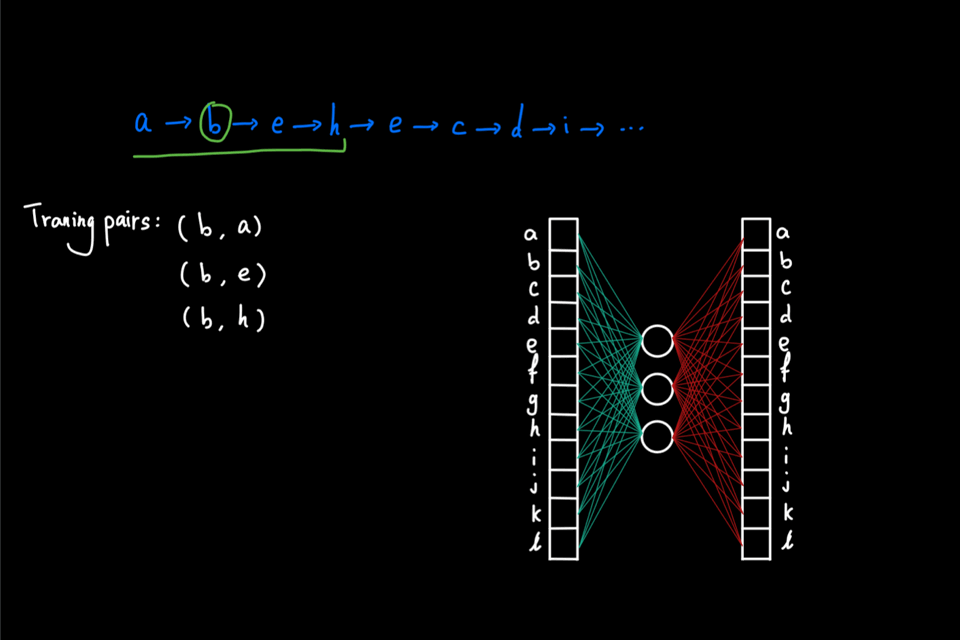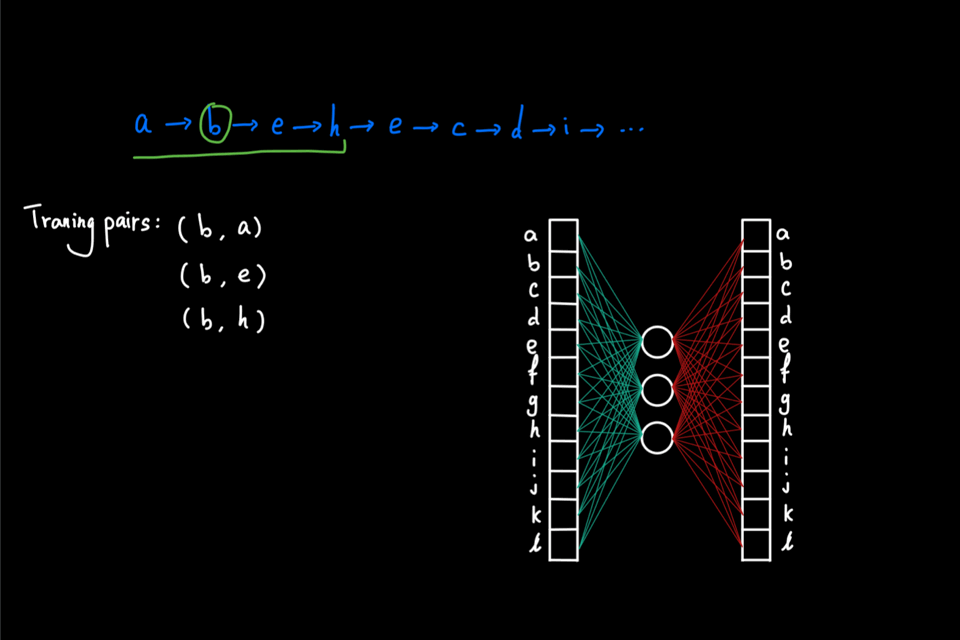
Why biased walk?
Now that we know how node2vec generates embedding using2nd order random walk on a graph, you might be wondering, why not just use 1st order random walk? The main reason for this is the flexibility of searching strategy. On the one hand, when the return parameter (p) and in-out parameter (q) are set to be 1, it recovers the first order random walk exactly. However, on the other hand, when p and q are set to be positive values other than 1, the random walks can be biased to either be localized in network modules or, conversely, be across the network.
Still, you might be wondering, why should you care if the walks are flexible in that sense?

The node2vec paper gave the following example of Les Miserables network embedding with different p and q. The resulting embeddings are used to perform clustering analysis to group nodes together. Nodes are then colored based on the cluster they belong to. The top and bottom panels correspond to the node2vec embedding generated using q = 0.5 and q = 2. One can see that in the top panel, nodes that fall into the same local network neighborhood (i.e., homophily) are colored the same. On the other hand, in the bottom panel, structurally equivalent nodes are colored the same. For example, the blue nodes correspond to characters that act as bridges between communities.
Closing remarks
Node2vec has been particularly popular in Computational Biology, mainly for gene classification using embeddings of Protein-Protein Interaction networks. For example, it has been shown that for gene function prediction, the PPI node2vec embedding followed by logistic regression can generate equal or better performance than the state-of-the-art network propagation methods [4]. Similarly, another method called GeneWalk uses node2vec to generate biological context-specific embeddings of genes and Gene Ontology terms [5].
Finally, despite the great performance of node2vec embeddings, the original implementations are not quite scalable due to memory issues. In particular, as we’ve seen before, node2vec devises 2nd order transition random walk, which depends on both the current state and the previous state. This causes the total number of transition probabilities to scale quadratically with respect to the number of edges in the graph (huge…). For example, it took over 100GB of memory to embed a decently sparse network with 17k nodes and 3.6M edges. This memory issue is effectively resolved by a new implementation called PecanPy, which only uses less than 1GB of memory to embed the same network [6]. You can also check out my other blog post for more information about PecanPy.
Thanks for your reading, and I hope this article gives you a better idea of how node2vec works!
Reference
[1] A. Grover, J. Leskovec, node2vec: Scalable Feature Learning for Networks (2016), ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD)
[2] C. McCormick, Word2Vec Tutorial – The Skip-Gram Model (2016)
[3] C. McCormick, Word2Vec Tutorial Part 2 – Negative Sampling (2017)
[4] Renming Liu, Christopher A Mancuso, Anna Yannakopoulos, Kayla A Johnson, Arjun Krishnan, Supervised learning is an accurate method for network-based gene classification (2020), Bioinformatics
[5] Ietswaart, R., Gyori, B.M., Bachman, J.A., et al. GeneWalk identifies relevant gene functions for a biological context using network representation learning (2021), Genome Biol
[6] R. Liu, A. Krishnan, PecanPy: a fast, efficient, and parallelized Python implementation of node2vec (2021), Bioinformatics







