Meta’s ESM-2 and ESMFold models for protein structure prediction
Two hallmarks of modern biology are (i) the irruption of ML models for predicting protein structures, resulting in a true revolution in the field, and (ii) the fact that this revolution was brought about by research labs from the private sector rather than by academics (luckily, these privates are leaving all code and models open therefore academics could further build on top of them).
The star and seed of the revolution I’m talking about was DeepMind with its Alphafold 2 model for protein structure prediction. Building on this, new ML models evolved, mainly in academic labs, that can perform protein design, prediction of interaction surfaces, and more:
New deep-learned tool designs novel proteins with high accuracy
New preprint describes a novel parameter-free geometric transformer of atomic coordinates to…
(For more, check this summary of all my articles on CASP, AlphaFold 2, and related technologies)
Then in the adjacent field of chemistry, I’ve presented how both DeepMind and Google are working on accelerating quantum calculations. Even TikTok seems to have plans to assist quantum calculations with ML methods, as it was recently hiring people formed in these areas.
DeepMind Strikes Back, Now Tackling Quantum Mechanical Calculations
Google proposes new method to derive analytical expressions for terms in quantum mechanics…
Meta, formerly Facebook, has been working for a couple of years on something quite new for biology, which is now bearing fruit: the development of a protein language model that "knows" about protein structure. As I will describe here, Meta created and evolved a series of methods that now ended up in a complete suite for protein structure prediction, design, and mutation evaluation, all fully based on language models.
As I explained in this recent article, the application of language models to protein structure prediction can help to overcome the limitation of tools like AlphaFold that rely on multiple sequence alignments, and also to accelerate dramatically the speed at which protein structures are predicted.
Recap of what protein structure modeling is, how it evolves, and the potential impact of protein language models
Very briefly, "protein structure modeling" is about predicting how proteins fold in 3D space from their amino acid sequences, and all kinds of related questions such as designing sequences of amino acids that fold into desired 3D structures. Both problems are central to biology: structure prediction for fundamental and applied biology, because scientists need to know protein structures in order to understand their functions and to design new drugs; and protein design for applied biotechnology for example to create new enzymes or stabilize existing ones.
Determining the 3D structures of proteins through experiments is very expensive and time-consuming, and might even remain unsuccessful after years of work. Hence the relevance of computational methods to predict 3D structures, ideally as accurately and as fast as possible. In fact, the problem of protein structure prediction is so central to biology and so hard to crack, that a competition between predictors is carried out every two years since 1994. The competition is called CASP, and was traditionally the arena of academics working on the problem. After years of no and then slow improvements, DeepMind cracked in CASP14 (2020) part of the protein structure prediction problem with their AlphaFold 2 program (AlphaFold 1 had previously won CASP13, but it was barely pushing to the limits everything that academics already knew, not really cracking the problem).
Now we get into AlphaFold’s limitations, and how language models could help. In order to model a protein from its sequence, AlphaFold 2 first builds an alignment of multiple sequences of proteins related to the query. This alignment is processed by a BERT-based language model specialized for proteins that then feeds numbers crunched from the alignment into the core of the network, which predicts the structure as its output.
The new methods developed by Meta (and also explored recently by some academics as I exemplified earlier) use language models much more advanced than BERT. These models learn so much about protein sequences and the evolutionary patterns that relate sequences to function, that then they don’t need sequence alignments at all in order to fold them. The latest such methods, just out from Meta and called ESM-2 and ESMFold, seem to predict protein structures as well as AlphaFold 2 but with a much faster prediction speed and without the need to compute alignments, as I explain and demonstrate in the next section.
But before proceeding to the explanations, let me highlight the relevance of Meta’s new methods by reminding the reader that AlphaFold 2’s results depend critically on the availability of large numbers of sequences with which to build a multiple sequence alignment. So, AlphaFold 2 is of limited utility for so-called "orphan" proteins, i.e. those proteins for which no alignments can be retrieved because not many sequences are available in databases. By not using alignments, methods based on language models might in principle model orphan proteins better than AlphaFold and similar methods, thus supposing a potential improvement in the reach of modern protein structure prediction -spoiler: the new methods help a bit, but not dramatically.
Additionally, by not requiring the compilation of an alignment, methods using language models run much faster than regular methods like AlphaFold, thus allowing the processing of larger numbers of sequences per unit of time -which Meta indeed exploited to process over 600 million sequences in just two weeks as I describe later on.
How Meta’s protein language models work to predict protein structures
This is fascinating. To develop its protein language models which culminated with ESM-2 and ESMFold, Meta experimented with training neural networks not to directly predict protein structures from a sequence (as in AlphaFold and other methods), but rather to predict the amino acids masked in protein sequences. That’s very similar to how language models like GPT-3 are trained: the algorithm masks tokens and the training procedure aims at predicting them. Somehow, ESM-2/ESMFold is "just another" huge language model but super specialized for proteins.
Like any other neural network, these protein language networks contain large numbers of weights (from millions to billions) that get finely tuned during training, in this case to predict masked residues. Meta discovered that when the network is well-trained to predict the masked amino acids in millions of natural protein sequences, then its internal weights are actually capturing, or "understanding", protein structure. See why I said this is fascinating?
Let me explain this again with different words. ESM-2’s training is only on sequences, both as input and output. Information about the structure being modeled develops within the network, as its weights describe the structural patterns that connect input (masked) sequences to output (complete) sequences. The protein structure is predicted "on the side" from the patterns activated inside the network as it processes the input sequence.
Somehow, it does make sense that in order to predict sequences, the network had to learn evolutionary patterns in the training sequences. And these patterns are well-known to relate directly to contacts between pairs of residues in protein structures -a concept well documented in CASP and structural Bioinformatics for almost a decade already, that made an impact on protein structure prediction already in CASP12 well before DeepMind entered the game with AlphaFold 1.
Going a bit deeper into detail, Meta already knew from its previous works that transformer models trained to model masked protein sequences develop attention patterns that correspond to the inter-residue contact map of the protein. What Meta did to teach ESM-2 how to actually derive a structure from its internal knowledge (thus creating ESMFold) was to project the attention patterns into known residue-residue contact maps obtained from experimental structures of the input sequences. Thus, when ESMFold processes an input sequence it immediately activates a series of attention patterns that it then converts into contact patterns which then are fed to a structure network that ends up computing the actual set of coordinates that conform the predicted structure.
How Meta arrived to ESM-2 and ESMFold
Meta’s works with proteins and language models started with what they presented in their 2019 paper in PNAS: evidence that language models trained on protein sequences internally learn protein properties related to structure and function. Then in 2020 Meta released ESM1b, a protein language model that already helped scientists to make some concrete predictions and discoveries about protein structure and function directly from protein sequences. Meta then scaled this up to create ESM-2, which at 15B parameters is the largest language model of proteins to date and makes the foundation for Meta’s modern tool for protein structure prediction and protein design. In developing ESM-2 and ESMFold, Meta observed that as the model was scaled up from 8 million to 15 billion parameters, sequence predictions get better and the information about protein structure extracted from the network’s attention patterns becomes richer and more accurate, thus effectively allowing to model protein structures by matching the weights to known structures.
Moreover, Meta found that protein structure predictions with ESM-2 are up to 60 times faster than with AlphaFold 2, reaching very similar accuracy (just a few points lower for some targets) without the need of any alignments, and slightly better results than AlphaFold 2 for orphan proteins -although not as dramatic as expectations were for language models.
Multiple ways to run ESMFold for structure prediction
In principle, Meta’s ESM-2 can be used to fold proteins, design proteins, and other tasks like predicting the effects of mutations on proteins. The main application released by Meta to date is ESMFold, a tool to fold proteins from their sequences, and an Atlas of precomputed models (covered in the next section).
Given a sequence, ESMFold outputs models and confidence metrics just like AlphaFold 2 does, i.e. the 1D pLDDT plot that estimates the accuracy with which each residue is modeled and the 2D PAE plot that estimates how well each residue is modeled relative to all the others.
Meta made ESMFold accessible in multiple ways. The simplest, right from their website, allows you to submit a sequence for modeling by using the "Fold sequence" functionality in this page:
The web service works very fast; for example I got this structure predicted in less than 4 seconds:

Notice that the model structure is colored to show the accuracy at each residue, i.e. by pLDDT. Blue means highly accurate and accuracy drops as you follow the colors of the rainbow until red is deemed probably wrong.
For richer structure prediction I recommend that you use the Google Colab notebook put together by sokrypton and colleagues, which provides full output including the much-needed metrics of model confidence in 1D (pLDDT) and 2D (PAE) (it does take a bit longer to run, though):
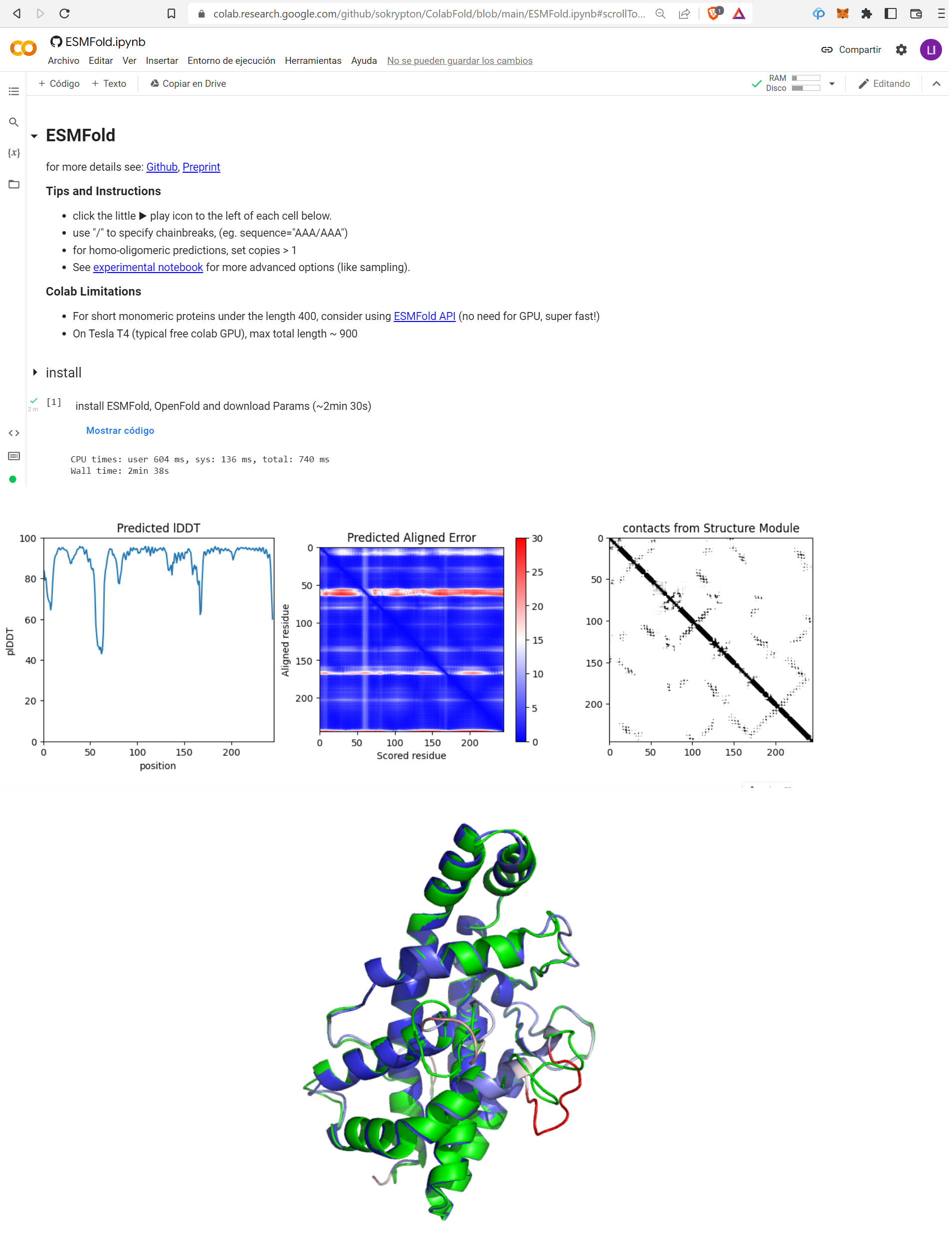
The panel at the bottom of this figure shows in green the actual structure as available in the PDB for this protein, compared to the ESMfold model colored by pLDDT (blue is high pLDDT i.e. high certainty, and red is low pLDDT i.e. uncertain). You can see that the only two unmatching parts are indeed predicted of low confidence by ESMfold (one somewhat off in pink and a very bad one in red).
Direct call to ESMFold through an API
When you use Meta’s service that I first presented above, you are actually accessing a very simple web page that sends your query sequence to an API that Meta put together to run the program.
This is clear in the format of the URL called when you submit a prediction:
https://esmatlas.com/resources/fold/result?fasta_header=Example&sequence=MSGMKKLYEYTVTTLDEFLEKLKEFILNTSKDKIYKLTITNPKLIKDIGKAIAKAAEIADVDPKEIEEMIKAVEENELTKLVITIEQTDDKYVIKVELENEDGLVHSFEIYFKNKEEMEKFLELLEKLISKLSGSGGGSGGGSGGGGSSGGGGGSGGGGSGGGGMSGMKKLYEYTVTTLDEFLEKLKEFILNTSKDKIYKLTITNPKLIKDIGKAIAKAAEIADVDPKEIEEMIKAVEENELTKLVITIEQTDDKYVIKVELENEDGLVHSFEIYFKNKEEMEKFLELLEKLISKLThis means that you can in principle make such a simple API call in any program or web app you are developing, and then process the obtained models right inside your own program!
A database of models for over 600 million proteins
Given how fast ESMFold runs, Meta could do something unprecedented in biology: they modeled 617 million proteins whose sequences were obtained from metagenomic projects, in just over 2 weeks! That’s unattainable even with AlphaFold 2, which is good at modeling but much slower than Meta’s new system.
Metagenomic projects entail sequencing the DNA of large numbers of organisms. But all that huge amount of information cannot be fully exploited without protein structures, or at least reliable models, hence the relevance of Meta’s new database of models—called ESM Metagenomic Atlas—in addition to the database of 200 million structures released by DeepMind together with the European Bioinformatics Institute.
Meta’s atlas of models can be browsed graphically as in the lead photo of this article, which is aesthetically appealing but doesn’t really have any utility. The true power of the Atlas is when coupled to its search engines: you can search the Atlas by MGnifyID (MGnify being a database of protein sequences from metagenomic datasets), by amino acid sequence (trying to find models already computed for that or similar sequences), or also by protein structure (to fish structurally similar models deposited in the database).
Closing words
When I thought the best of protein structure prediction had happened (with AlphaFold 2), Meta came out with this fascinating approach, tool, and database. With the scientific abstracts for CASP15 just out and without news from DeepMind but with news from Meta, I wonder what surprises we may have if any. On one side preliminary evaluations seem to show no big improvements relative to CASP14, but on the other hand AlphaFold 2 models were already so good that there is little space for improvement, and as I discussed CASP15 is moving towards new goals. Prediction of orphan proteins could be impacted at least a bit by Meta’s ESMFold, but such proteins usually don’t abound in CASP. We will know soon, just 3 weeks when CASP15 results are released, if language models (not only Meta’s but also others being developed by academics) can further foster the revolution.
References
Preprint of the main ESM-2 network, ESMFold, and the ESM atlas:
Evolutionary-scale prediction of atomic level protein structure with a language model
The main website making ESM-2 tools and Atlas available:
Earlier related works on how language models help predict the effects of mutations in proteins, and to design proteins:
Language models enable zero-shot prediction of the effects of mutations on protein function
Learning inverse folding from millions of predicted structures
www.lucianoabriata.com I write and photoshoot about everything that lies in my broad sphere of interests: nature, science, technology, programming, etc. Become a Medium member to access all its stories (affiliate links of the platform for which I get small revenues without cost to you) and subscribe to get my new stories by email. To consult about small jobs check my services page here. You can contact me here.







